Research Projects
Identifying protein kinase C-β-dependent mechanisms of TME-remodelling
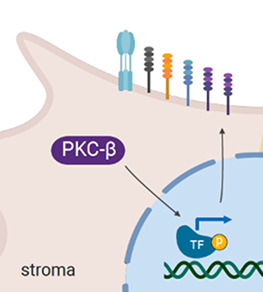
Our lab has identified that cancer cells derived from different tissues can induce the expression of PKC-β, which orchestrates gene expression through the control of transcription factors (TF) in mesenchymal cells of the TME. Inhibiting protein expression or antagonising the kinase activity of PKC-β significantly impairs disease progression in in vivo models of B cell leukaemia and some solid cancer models. We are now seeking to identifying the mechanisms underlying the tumour-promoting effects of PKC-β expressed in cells of the TME. A focus of our work lies on understanding how stromal PKC-β regulates plasma membrane stability and how this effects tumour cell intrinsic signalling and apoptosis control.
Papers: Lutzny et al Cancer Cell 2013; Park et al Science Translational Medicine 2020
Characterising the nexus between clonal evolution, stroma reprogramming and drug-responses
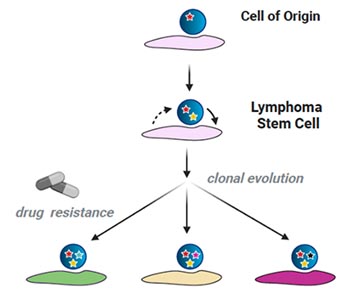
While targeted therapies have substantially improved the outcome of many cancer patients, the vast majority of patients cannot be cured and palliation remains the treatment goal. Genomic instability and clonal evolution have been recognised to be the main driver of acquired drug resistance. We and others have shown that interactions with the TME can protect cancer cells from cancer therapies. Our current work focuses on identifying oncogene-specific reprogramming of the TME and to understand how this impacts on responses to treatment.
Papers: Mangolini et al. Nature communications 2022
Dissecting spatially-defined functions of the
tyrosine kinase ZAP-70
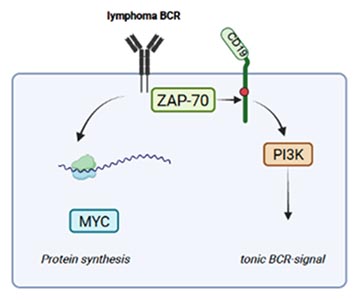
The tyrosine-kinase ZAP-70 is essential for T cell receptor signalling, but aberrantly expressed in a subset of B cell cancers. In tumour B cells it can augment a tonic, constitutive B cell receptor signal, which has implications for gene expression, T cell support, migration, MYC-expression and protein synthesis. Evidence from our group indicates that these divers functions of ZAP-70 are regulated through spatially defined protein-protein interactions. We are aiming to dissect these functions to understand mechanisms of B and T cell activation.
Papers: Chen et al, Blood 2020 and Blood advances 2024